---
tags:
- antibody
- antigen
- binding
- COVID-19
- SARS-CoV-2
- benchmark
pretty_name: CoV-UniBind
---
CoV-UniBind - Coronavirus Unified Binding Database
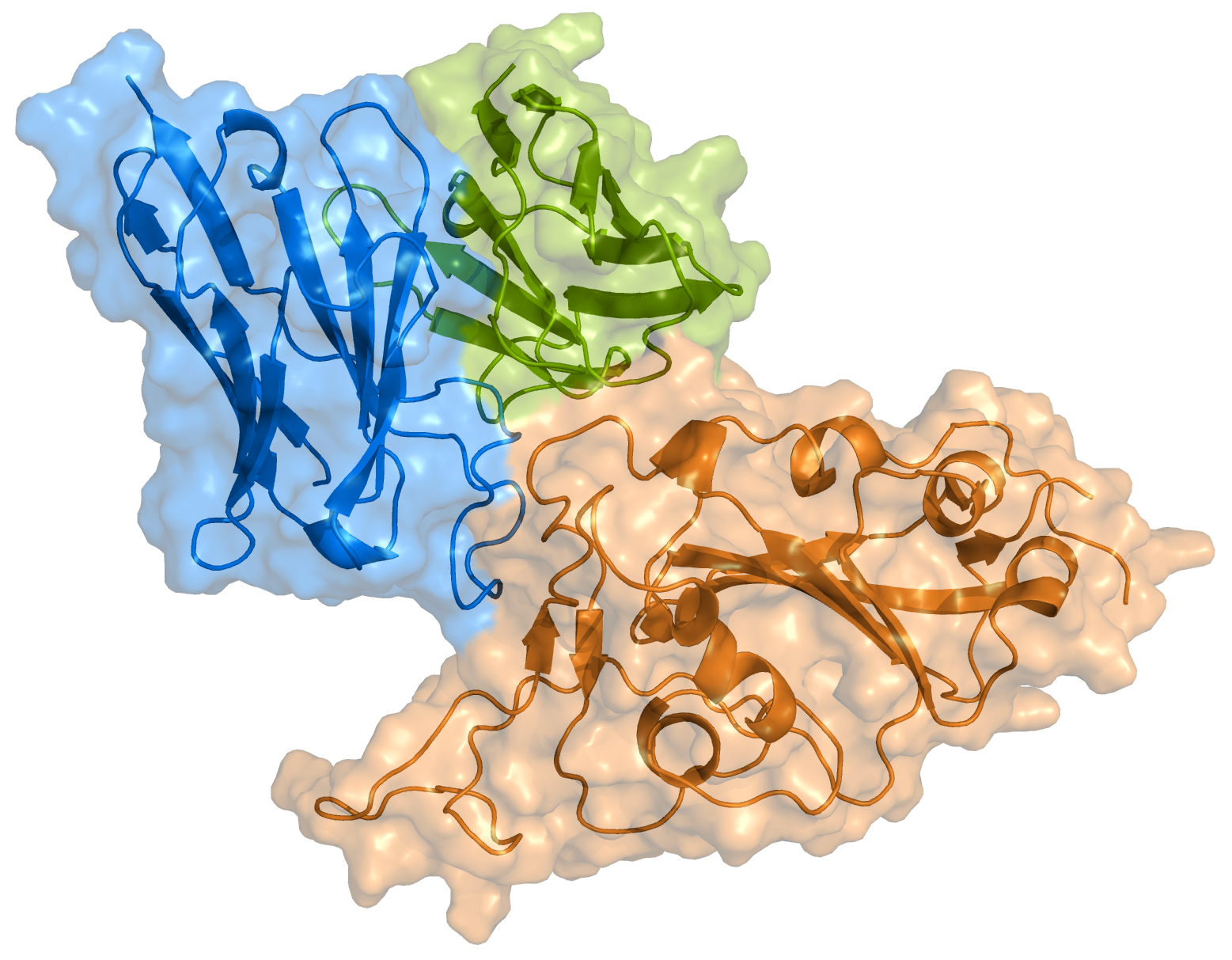
**CoV-UniBind** curates and integrates structural and biochemical data on antibodies specifically elicited by SARS-CoV-2 and
related coronaviruses. It links 3D antibody structures to their binding properties across viral variants, incorporating
epitope and sequence information. This dataset serves as a comprehensive resource for analysing antibody-antigen
interactions in the context of COVID-19 and provides a foundation for binding classification and regression tasks using
deep learning models.
## Database Overview
| Dataset | Description | Label | Type | References (doi) |
|------|-------------|-------|-----| ------- |
| covabdab | Binding labels from the Coronavirus Antibody Database (CoV-AbDab) | binding | bool | 10.1093/bioinformatics/btaa739 |
| dms_bloom | Deep Mutational Scanning escape data from Greaney et al. 2022 | escape | float | 10.1038/s41564-021-00972-2; 10.1016/j.chom.2020.11.007; 10.1038/s41467-021-24435-8; 10.1016/j.xcrm.2021.100255; 10.1126/science.abf9302; 10.1038/s41467-021-24435-8; 10.1038/s41564-021-00972-2; 10.1038/s41586-021-04385-3; 10.1038/s41586-022-04980-y; 10.1038/s41586-021-03817-4; 10.1038/s41586-021-03807-6 |
| dms_cao | Deep Mutational Scanning escape data from Cao et al. 2023 | escape | float | 10.1038/s41586-022-05644-7 |
| jian_elisa | ELISA antibody IC50 measurements from Jian et al. 2025 | IC50 | float | 10.1038/s41586-024-08315-x |
| spr | Surface Plasmon Resonance antibody affinity measurements from multiple sources | KD | float | 10.1038/s41467-024-54916-5; 10.1038/s41586-020-2380-z; 10.1038/s41467-021-24514-w; 10.1016/j.immuni.2022.06.005; 10.1016/j.xcrm.2023.100991 |
| drdb | Neutralisation potency data from the SARS-CoV-2 Resistance Database (DRDB) | IC50 | float | 10.1371/journal.pone.0261045 |
## Database Structure
```
.
├── antibody_info
│ └── antibody_synonyms.csv
├── data
│ ├── covabdab_binding.parquet
│ │ └── structures
│ │ ├── processed.zip
│ │ └── trimmed.zip
│ ├── dms_bloom_ab_escape.parquet
│ ├── dms_cao_ab_escape.parquet
│ ├── drdb_binding_potency.parquet
│ ├── jian_elisa_ab_ic50.parquet
│ └── spr_ab_affinity.parquet
├── scores
│ ├── covabdab_binding_scores.parquet
│ ├── dms_bloom_ab_escape_scores.parquet
│ ├── dms_cao_ab_escape_scores.parquet
│ ├── drdb_binding_potency_scores.parquet
│ ├── jian_elisa_ab_ic50_scores.parquet
│ └── spr_ab_affinity_scores.parquet
├── cov-unibind.py
└── README.md
```
## Usage Guide
```python
from datasets import load_dataset
data='drbd' # specify the dataset name based on table above
dataset = load_dataset("InstaDeepAI/cov-unibind",name=data)
```
## Dataset Schema
The table below includes information about the columns contained in the datasets.
| Column Name | Description | Type | Nullable | Example |
|---|---|---|---|---|
| `antibody_name` | Name of the antibody | *str* | False | `bd30_515;bd_515` |
| `antigen_lineage` | Antigen lineage | *str* | False | `BA.1` |
| `target_value` | Experimental binding value| *float* or *bool* | False | `-2.327902` or `True` |
| `target_type` | Type of target value | *str* | False | `IC50_log10_fold` |
| `source_name` | Source of the data | *str* | False | `jian_2024_nature` |
| `source_doi` | DOI of the source | *str* | False | `10.1038/s41586-024-08315-x` |
| `assay_name` | Name of the assay | *str* | False | `elisa` |
| `pdb_id` | PDB structure ID | *str* | False | `7e88` |
| `structure_release_date` | Release date of the structure | *str* | False | `03/01/21` |
| `structure_resolution` | Resolution of the structure (Å) | *float* | False | `3.14` |
| `mutations` | Lineage consensus mutations | *str* | False | `A67V H69- V70- T95I...` |
| `antigen_chain_ids` | Chain IDs of the antigen | *str* | False | `C` |
| `antigen_domain` | Domain of the antigen | *str* | False | `RBD` |
| `antigen_residue_indices` | Residue indices of the antigen | *str* | False | `(13, 568)` |
| `antigen_residue_indices_trimmed` | Antigen residue indices, trimmed | *float* | True | `(333, 526)` |
| `antigen_host` | Host organism of the antigen | *str* | False | `severe acute respiratory syndrome coronavirus 2 (2697049)` |
| `antibody_heavy_chain_id` | Heavy chain ID of the antibody | *str* | False | `C` |
| `antibody_light_chain_id` | Light chain ID of the antibody | *str* | False | `B` |
| `epitope_residues` | Residues of the epitope | *str* | False | `R403 D405 T415` |
| `epitope_mutations` | PDB antigen mutations in the epitope | *str* | True | `D405T` |
| `epitope_domain` | Spike domain where the antibody binds | *str* | False | `RBD` |
| `epitope_alteration_count` | Number of alterations in the epitope | *float* | True | `2` |
| `spike_sequence` | Full spike protein sequence | *str* | False | `MFVFLVLLPLVSSQCVNLTTRTQL...` |
| `antibody_heavy_chain_sequence` | Sequence of the antibody heavy chain | *str* | False | `EVQLVESGGGLVQPGGSLRLSCAASEFIVSRNYMSWVR...` |
| `antibody_light_chain_sequence` | Sequence of the antibody light chain | *str* | False | `DIQMTQSPSSLSASVGDRVTITCQASQDINKYLNWYQQK...` |
| `antibody_vh_sequence` | Sequence of the VH domain | *str* | False | `EVQLVESGGGLVQPGGSLRLSCAASEFIVSRNYMSWVRQ...` |
| `antibody_vl_sequence` | Sequence of the VL domain | *str* | False | `DIQMTQSPSSLSASVGDRVTITCQASQDINKYLNW...` |
| `antigen_sequence` | Full antigen sequence | *str* | False | `TNLCPFDEVF-NATRFASVYAWNR--KRISNCVADYSVLYNLAPFFTFKCYGVSP...` |
| `antigen_sequence_trimmed` | Trimmed antigen sequence | *float* | True | `TRFASV-YAWNRKRISNCVADYSVLYNLAPFFT-FKCYGVSP...` |
| `antigen_sequence_without_indels`| Antigen sequence without insertions/deletions | *str* | False | `TNLCPFDEVFNATRFASVYAWNRKRISNCVAD...` |
| `antigen_sequence_trimmed_without_indels` | Trimmed antigen sequence without insertions/deletions | *float* | True | `NATRFASVYAWNRKRISNCVAD...` |
| `antigen_pdb_sequence` | Antigen sequence from PDB | *str* | False | `TNLCPFGEVFNATRFASVYAWNRKRISNCVADYSVLYNSA...` |
| `antigen_pdb_sequence_trimmed` | Trimmed antigen sequence from PDB | *float* | True | `NATRFASVYAWNRKRISNCVADYSVLYNSA...` |
## Acknowledgments
This project makes use of publicly available antibody datasets listed above. We acknowledge the contributions by the teams
responsible for compiling and maintaining these valuable resources.

